ATCC-Center for Translational Microbiology will present at the Society for Industrial Microbiology (SIMB) Annual Meeting in Denver, CO (July 29-August 3). They will be presenting research of the development of synthetic and uniquely tagged whole cells as spike-in controls for microbiome research applications.
Title: Validation of Differentiable Molecular Barcodes as Reference Standards for Microbiome Analyses
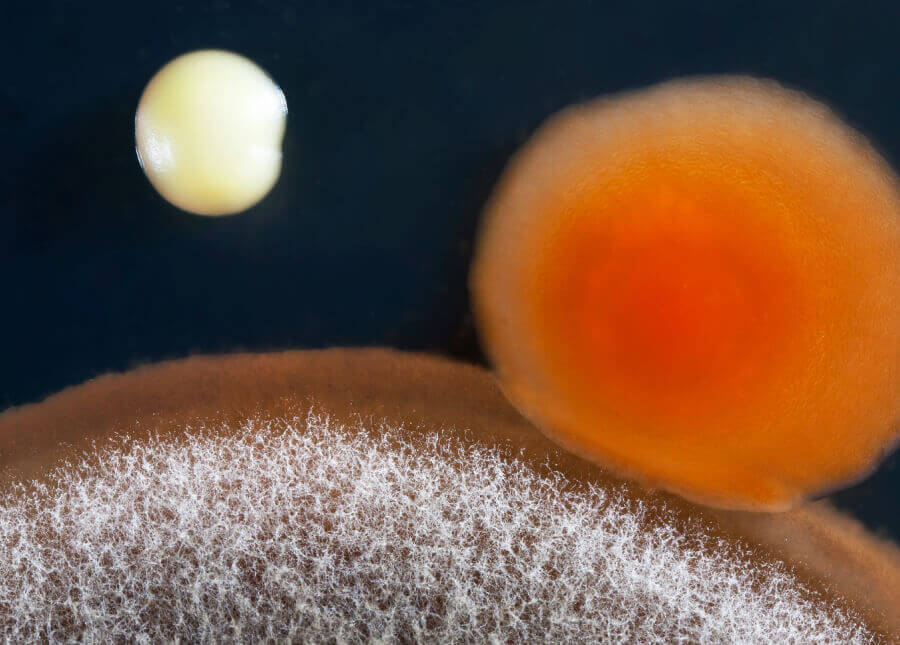
Abstract: A shortcoming in microbiome studies is the lack of reference standards to control biases introduced by DNA extractability, library preparation, and sequencing, which can limit the accuracy of data interpretation. To address this, five differentiable molecular barcodes were spiked in staggered copy numbers (106 and 105) into porcine fecal or DNA samples as oligonucleotides, transformants or recombinants, and quantitated using bioinformatics tools. Additionally, Escherichia coli and Lactobacillus gasseri recombinants and transformants with two different barcoded tags were also spiked at copy numbers of 106 and 105, respectively. DNA was extracted three commercially available kits. 16S rRNA gene high-throughput sequencing and shotgun metagenomics were performed using Illumina MiSeq to determine the level of detectability of the molecular barcodes using bioinformatic analyses. Results showed that mapping of 16S rRNA gene and shotgun metagenomic data against each of the differentiable molecular barcodes as the reference sequences provided a high level of detectability and quantification. Spiking plasmids containing the differentiable molecular barcodes resulted in lower recovered copies (102-103) compared to E. coli (106) and L. gasseri (101) spiked transformants, possibly due to loss during DNA extraction, library preparation and/or sequencing. Operational Taxonomic Unit (OTU) picking strategy had an effect in diversity and taxonomy analyses. Results support the value of including reference standards in microbiome and metagenome analyses to strengthen the validity of findings in microbiome research.
For further information or to contact ILSE scientist at the event email: [email protected]